We are delighted to celebrate an important achievement within our research community: the publication of the PheWeb2 brief paper in Nature Genetics. Congratulations to Hongyu Xiao and Daniel Taliun, as well as their collaborators, for this outstanding accomplishment!
Their article, “Exploring and visualizing stratified GWAS results with PheWeb2”, was published on 18 January 2026 in Nature Genetics (DOI: 10.1038/s41588-025-02469-8).
Advancing the Exploration of Stratified GWAS Data
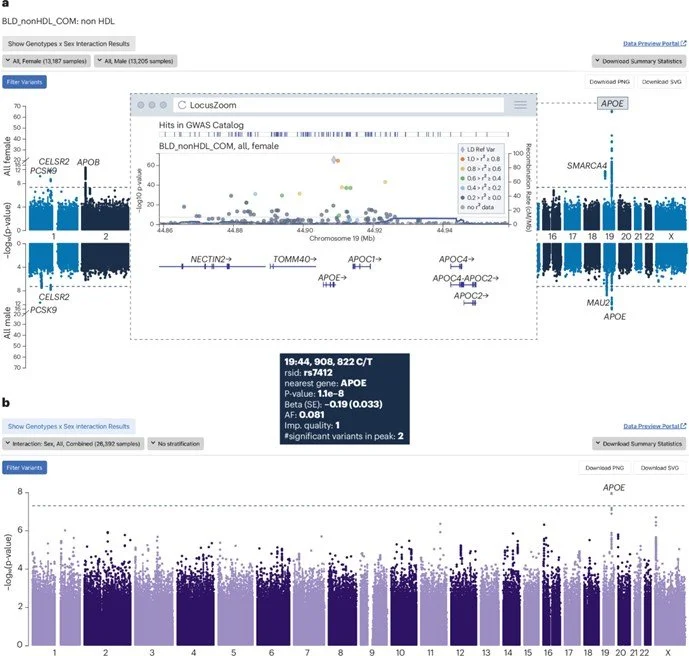
PheWeb2 represents a complete redesign and enhancement of the original PheWeb tool, long used by the genetics community to navigate and share large-scale genome‑wide association study (GWAS) results. The new version addresses a major bottleneck in current research: the lack of intuitive tools for exploring sex‑stratified, ancestry‑stratified, and other stratified GWAS summary statistics.
The redesigned platform introduces major improvements, including:
Interactive visual comparisons such as Miami plots and stacked LocusZoom plots for side‑by‑side analysis of stratified GWAS or PheWAS results.
Support for numerous types of stratification, including genetic ancestry groups and sex‑specific analyses.
A modernized software architecture, decoupling the API from the user interface to improve maintainability and facilitate integration with external tools.
A fully updated UI built with Vue 3 and Vite, enhancing performance and ease of feature development.
These advances make PheWeb2 a powerful tool for researchers aiming to uncover subtle genetic effects that vary between population groups, ultimately helping drive new biological insights from increasingly diverse genomic datasets.
Please join us in congratulating Hongyu and Daniel on this exciting achievement!